We work with our customers in precision medicine to solve problems in engineering, bioinformatics, science, and translational medicine that involve clinical and omics data.
We work with our customers in precision medicine to solve problems in engineering, bioinformatics, science, and translational medicine that involve clinical and omics data.
Genomics Shaping Healthcare
Genomics Shaping Healthcare

Deep First Hand Experience
We have deep first-hand experience in the area, spanning tens of thousands of reports.
Technology
Our fast and nearly automated platform for small and large oncology panel reporting is based on ~10 years curation and automation.
Customer Experience
Our teams have helped build germline and somatic variant interpretation tools for multiple US diagnostic and device companies.
Data Generation
We generate single-cell, spatial transcriptomics and genomics data from clinical samples in our India-based CAP lab.
Data Curation
We curate publicly available and client-specific omics data and develop custom ontologies for metadata and cell type annotations, with focus on single-cell and spatial transcriptomics.
Data Analysis
We build bioinformatics and tertiary analysis workflows that help go from raw reads to insights, and package these in software platforms to help lab scientists, bioinformaticians, and curation scientists.
Track Record
We have a 20-year strong track record in product development for omics applications, e.g., GeneSpring, MassProfiler Professional, and StrandNGS.
Wide Range of Experience
We have experience building visualization tools, rules engines, data management systems, user interfaces, and algorithms. We have built applications for microarrays, genomics, transcriptomics, metabolomics, and proteomics, as well as for metadata management, clinical reporting, electronic data capture, and provider portals.
Software Development for
Regulatory Compliance
We develop software for HIPAA- and FDA-compliant software accompanying medical devices and genomics tests.
Excellence Through the Years
Excellence Through the Years
Drawing on 24 years of domain experience, we are a team of over 300 highly trained curation scientists, software engineers, testing engineers, bioinformaticians, and clinical researchers.
We are headquartered in Bangalore, which is also where we were founded, and have close ties with the Bay Area and San Diego, where many of our customers are based.
Quality
Quality is a core tenet of the work delivered at Strand. Our team of over 500 helps ensure that everything we do meets the highest standards through rigorous internal checks and validation.
Innovation
Our team of ~ 340 specialists adapts to and innovates on breakthroughs in the ever-evolving landscape of precision omics using cutting-edge technologies including AI-based solutions, RWD-derived results, and multiomics analysis.
Collaboration
We work closely with clients across academic, pharmaceutical, bioinformatics, biotechnology, and software companies; providing solutions in data harmonization, precision analytics, software tool and platform development, and quality control, to name a few.
Global Perspective
With compliance to internationally recognized standards such as HIPAA, and ISO 20071, coupled with the CAP accreditation for our NGS laboratory, our global team serves clients worldwide.
Testimonials

15 Dec 2025
Written by Suhasini Singh
26 Nov 2025
Written by Sharon Christella

17 Nov 2025
Written by Sharon Christella
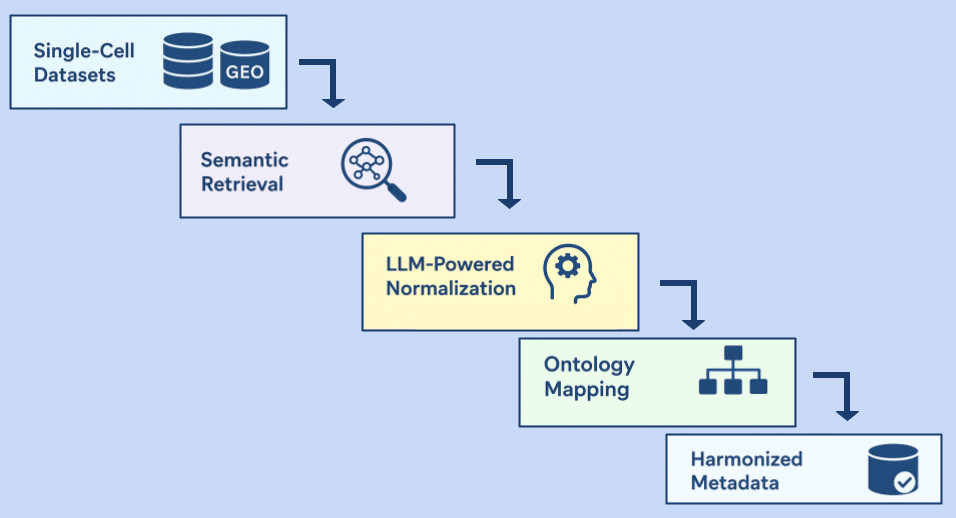
11 Sep 2025
Written by Badri Padhukasahasram, Shrutee Jakhanwal and Chinta Sidharthan

30 Jun 2025
Written by Suhasini Singh

26 Jun 2025
Written by Sharon Christella
Insights from our CEO
Insights from our CEO
Leaders in Genomics